|
Examples / CVMODEL.RPF |
CVMODEL.RPF is an example of Structural VAR estimation. It estimates two structural models for the six-variable Canadian VAR used in IMPULSE.RPF as well as several other examples. It demonstrates the use of the CVMODEL and ERRORS instructions.
The first model is a “B” type model which interprets the orthogonal shocks as two real shocks, two financial market shocks and two nominal shocks. The loadings of these shocks onto the innovations produces a highly overidentified model: there are only six free parameters instead of the fifteen for a just identified model.
\begin{equation} \left[ {\begin{array}{*{20}c} {u_u } \\ {u_c } \\ {u_r } \\ {u_x } \\ {u_m } \\ {u_p } \\ \end{array}} \right] = \left[ {\begin{array}{*{20}c} 1 & 0 & {\beta _{uf1} } & 0 & 0 & 0 \\ {\beta _{cr1} } & 1 & {\beta _{cf1} } & 0 & 0 & 0 \\ 0 & 0 & 1 & {\beta _{rf2} } & 0 & 0 \\ 0 & 0 & 0 & 1 & 0 & 0 \\ {\beta _{mr1} } & 0 & 0 & 0 & 1 & {\beta _{mn1} } \\ 0 & 0 & 0 & 0 & 0 & 1 \\ \end{array}} \right]{\kern 1pt} {\kern 1pt} {\kern 1pt} {\kern 1pt} \left[ {\begin{array}{*{20}c} {v_{r1} } \\ {v_{r2} } \\ {v_{f1} } \\ {v_{f2} } \\ {v_{n1} } \\ {v_{n2} } \\ \end{array}} \right] \end{equation}
The second model is also overidentified, with nine parameters. This is an “A” type model, where non-orthogonalized shocks are mapped to orthogonalized ones.
\begin{equation} \left[ {\begin{array}{*{20}c} 1 & 0 & {\gamma _{ur} } & 0 & 0 & 0 \\ {\gamma _{cu} } & 1 & {\gamma _{cu} } & 0 & 0 & 0 \\ 0 & 0 & 1 & {\gamma _{rx} } & 0 & 0 \\ 0 & 0 & 0 & 1 & 0 & 0 \\ 0 & {\gamma _{mc} } & {\gamma _{mr} } & 0 & 1 & {\gamma _{mp} } \\ 0 & {\gamma _{pc} } & {\gamma _{pr} } & 0 & 0 & 1 \\ \end{array}} \right]{\kern 1pt} {\kern 1pt} {\kern 1pt} {\kern 1pt} \left[ {\begin{array}{*{20}c} {u_u } \\ {u_c } \\ {u_r } \\ {u_x } \\ {u_m } \\ {u_p } \\ \end{array}} \right] = \left[ {\begin{array}{*{20}c} {v_u } \\ {v_c } \\ {v_r } \\ {v_x } \\ {v_{_m } } \\ {v_p } \\ \end{array}} \right] \end{equation}
The setup of the basic VAR is the same as for IMPULSE.RPF, though with a slightly different order on the variables. (You can use any listing order for a structural model, through you have to make sure you define the model based upon the order you choose). These are the initial declarations for the first structural VAR. We need a FRML which evaluations to a RECTANGULAR matrix, which needs to be declared before it can be created. We also need to declare the free parameters:
dec frml[rect] bfrml
nonlin uf1 cr1 cf1 rf2 mf1 mm2
In this case, we’re using “descriptive” names for these (for instance, UF1 is for the loading of the F1 shock onto US GDP, CR1 for the loading of R1 onto the Canadian GDP). You could also use positions like B13, B21, etc. The following constructs the FRML describing the structural model. We strongly recommend using one row per line (with the $ continuation) and using spaces or zero padding to make the columns line up. It will save you a lot of time if you have to make changes, or have some problem with the model. In this case, all parameters are initialized to zero, which often works OK for a model like this. The rows correspond to the dependent variables in the order in which you put them into the model, while the columns correspond to your intended shocks.
frml bfrml = ||1.0,0.0,uf1,0.0,0.0,0.0|$
cr1,1.0,cf1,0.0,0.0,0.0|$
0.0,0.0,1.0,rf2,0.0,0.0|$
0.0,0.0,0.0,1.0,0.0,0.0|$
mf1,0.0,0.0,0.0,1.0,mm2|$
0.0,0.0,0.0,0.0,0.0,1.0||
compute uf1=cr1=cf1=rf2=mf1=mm2=pm2=0.0
This is estimated by using the genetic annealing method first, then polishing the estimates with BFGS. (Structural VAR's can have multiple modes, hence the use of the slower, broader search for the PMETHOD).
cvmodel(b=bfrml,method=bfgs,factor=bfactor,$
pmethod=ga,piters=20) %sigma
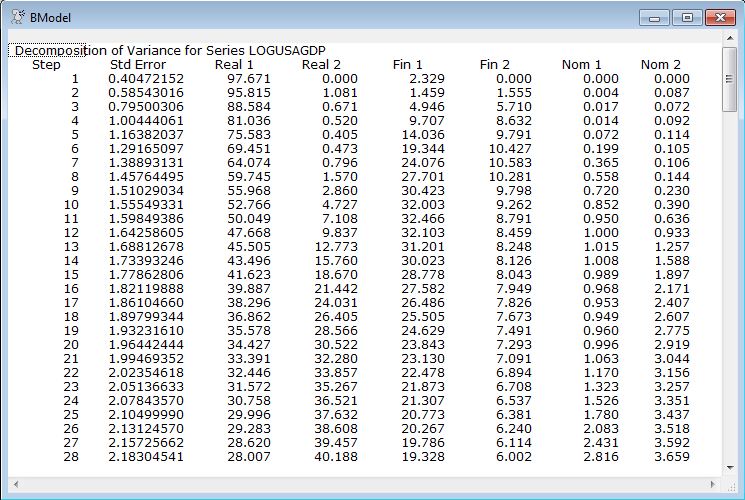
Because the shocks don’t really correspond one-to-one with the variables, the LABELS option is used on ERRORS to give them the desired labels.
errors(model=canmodel,factor=bfactor,steps=28,window="BModel",$
labels=||"Real 1","Real 2","Fin 1","Fin 2","Nom 1","Nom 2"||)
The same procedure is followed with the other model. It just uses the A option on CVMODEL rather than the B option:
dec frml[rect] afrml
nonlin rx ur cu cr pc pr mp mc mr
frml afrml = ||1.0,0.0,ur ,0.0,0.0,0.0|$
cu ,1.0,cr ,0.0,0.0,0.0|$
0.0,0.0,1.0,rx ,0.0,0.0|$
0.0,0.0,0.0,1.0,0.0,0.0|$
0.0,mc ,mr ,0.0,1.0,mp |$
0.0,pc ,pr ,0.0,0.0,1.0||
compute ur=cu=cr=rx=mc=mr=mp=pc=pr=0.0
cvmodel(a=afrml,method=bfgs,factor=afactor,$
pmethod=ga,piters=20) %sigma
errors(model=canmodel,factor=afactor,steps=28,window="AModel")
Full Program
open data oecdsample.rat
calendar(q) 1981
data(format=rats) 1981:1 2006:4 can3mthpcp canexpgdpchs $
canexpgdpds canm1s canusxsr usaexpgdpch
*
set logcangdp = 100.0*log(canexpgdpchs)
set logcandefl = 100.0*log(canexpgdpds)
set logcanm1 = 100.0*log(canm1s)
set logusagdp = 100.0*log(usaexpgdpch)
set logexrate = 100.0*log(canusxsr)
*
system(model=canmodel)
variables logusagdp logcangdp can3mthpcp logexrate logcandefl logcanm1
lags 1 to 4
det constant
end(system)
*
estimate(noprint)
*
dec frml[rect] bfrml
nonlin uf1 cr1 cf1 rf2 mf1 mm2
frml bfrml = ||1.0,0.0,uf1,0.0,0.0,0.0|$
cr1,1.0,cf1,0.0,0.0,0.0|$
0.0,0.0,1.0,rf2,0.0,0.0|$
0.0,0.0,0.0,1.0,0.0,0.0|$
mf1,0.0,0.0,0.0,1.0,mm2|$
0.0,0.0,0.0,0.0,0.0,1.0||
compute uf1=cr1=cf1=rf2=mf1=mm2=pm2=0.0
*
* This is estimated by using the genetic method first, then polishing the
* estimates with bfgs. In practice, you might want to repeat this
* several times to test whether there are global identification problems.
*
cvmodel(b=bfrml,method=bfgs,factor=bfactor,$
pmethod=ga,piters=20) %sigma
*
* Because the shocks don't really correspond one-to-one with the
* variables, the labels option is used on ERRORS to give them the
* desired labels.
*
errors(model=canmodel,factor=bfactor,steps=28,window="BModel",$
labels=||"Real 1","Real 2","Fin 1","Fin 2","Nom 1","Nom 2"||)
dec frml[rect] afrml
nonlin rx ur cu cr pc pr mp mc mr
frml afrml = ||1.0,0.0,ur ,0.0,0.0,0.0|$
cu ,1.0,cr ,0.0,0.0,0.0|$
0.0,0.0,1.0,rx ,0.0,0.0|$
0.0,0.0,0.0,1.0,0.0,0.0|$
0.0,mc ,mr ,0.0,1.0,mp |$
0.0,pc ,pr ,0.0,0.0,1.0||
compute ur=cu=cr=rx=mc=mr=mp=pc=pr=0.0
cvmodel(a=afrml,method=bfgs,factor=afactor,$
pmethod=ga,piters=20) %sigma
errors(model=canmodel,factor=afactor,steps=28,window="AModel")
Output
Covariance Model-Concentrated Likelihood - Estimation by BFGS
Convergence in 14 Iterations. Final criterion was 0.0000004 <= 0.0000100
Observations 100
Log Likelihood -585.4719
Log Likelihood Unrestricted -578.6729
Chi-Squared(9) 13.5981
Significance Level 0.1373572
Variable Coeff Std Error T-Stat Signif
************************************************************************************
1. UF1 0.102630163 0.070065484 1.46477 0.14298234
2. CR1 0.500515182 0.104290402 4.79924 0.00000159
3. CF1 -0.077813852 0.079754083 -0.97567 0.32922687
4. RF2 -0.084722322 0.036190823 -2.34099 0.01923273
5. MF1 -0.161403447 0.108387565 -1.48913 0.13645248
6. MM2 0.112152267 0.042611628 2.63196 0.00848929
Covariance Model-Concentrated Likelihood - Estimation by BFGS
Convergence in 18 Iterations. Final criterion was 0.0000028 <= 0.0000100
Observations 100
Log Likelihood -581.0251
Log Likelihood Unrestricted -578.6729
Chi-Squared(6) 4.7044
Significance Level 0.5822397
Variable Coeff Std Error T-Stat Signif
************************************************************************************
1. RX 0.096444096 0.036861996 2.61636 0.00888739
2. UR -0.099391995 0.072913270 -1.36315 0.17283407
3. CU -0.504941212 0.104916628 -4.81279 0.00000149
4. CR 0.146654170 0.078837808 1.86020 0.06285709
5. PC -0.226893636 0.227477992 -0.99743 0.31855528
6. PR 0.306165446 0.165721688 1.84747 0.06467940
7. MP -0.121270363 0.048212171 -2.51535 0.01189151
8. MC 0.204902146 0.105038542 1.95073 0.05108882
9. MR 0.040661738 0.086947863 0.46766 0.64003022
Reports
This generates two rather lengthy reports with the error decompositions from the two models. Below is part of one of those (for the second "B" model).
Copyright © 2026 Thomas A. Doan